- Share
- Share on Facebook
- Share on X
- Share on LinkedIn
Identification and characterization of small regulatory RNAs and transcription attenuators in E. coli
Stéphan Lacour, Alexandre Dawid
Identification of small regulatory RNAs in E. coli
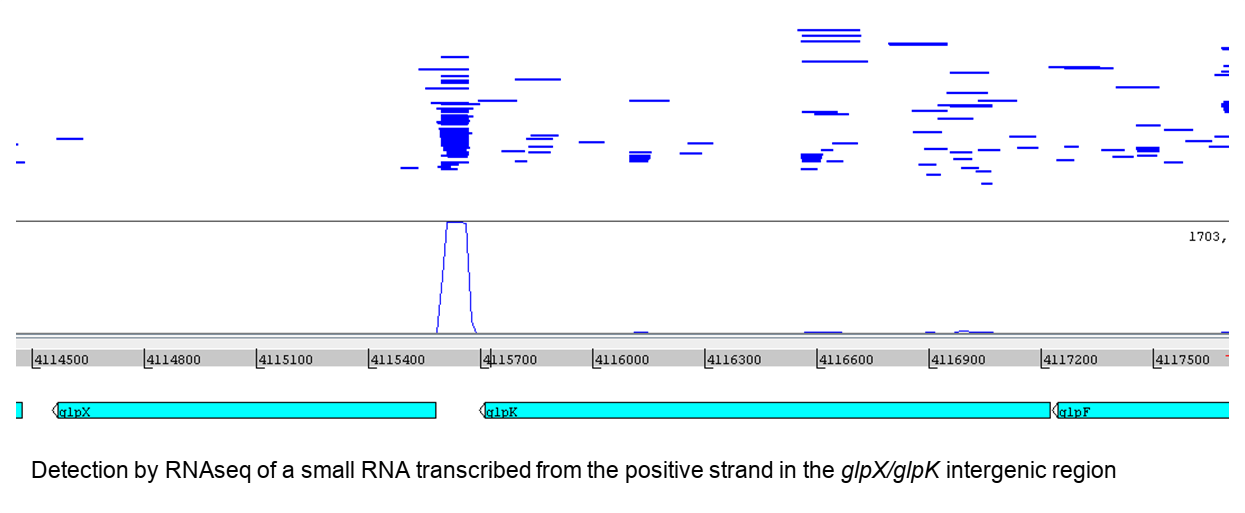
Using data from massive sequencing of small transcripts in Escherichia coli, we have identified numerous small RNAs transcribed independently of protein-coding genes. The role of some of them could be to regulate gene expression by acting in trans on the stability and translation rate of different messenger RNAs. We aim to validate this hypothesis experimentally, using bioinformatics tools to predict potential gene targets and experimental data on RNA-RNA interactomes (with immunoprecipitation of a chaperone RNA). We are focusing in particular on candidate non-coding RNAs from intergenic regions. We will then extend the study to detections from UTR (untranslated regions) flanking protein-coding sequences. After experimental validation of these detections (Northern, RT-PCR, other), we look for their gene targets (bioinformatic predictions or published interactomes) and set up experiments to demonstrate the regulation of one or more biological functions by these new small RNAs.
Identification of new transcription attenuators in E. coli
In parallel with the search for new small trans-regulatory RNAs, we are also seeking to identify transcription attenuators in the 5'UTR regions we have identified as sources of small RNAs. Indeed, some of these small RNAs originating from 5'UTR regions could be the result of premature transcription termination.
- Share
- Share on Facebook
- Share on X
- Share on LinkedIn