- Share
- Share on Facebook
- Share on X
- Share on LinkedIn
Design and dynamics of RNA folding
Alexandre Dawid
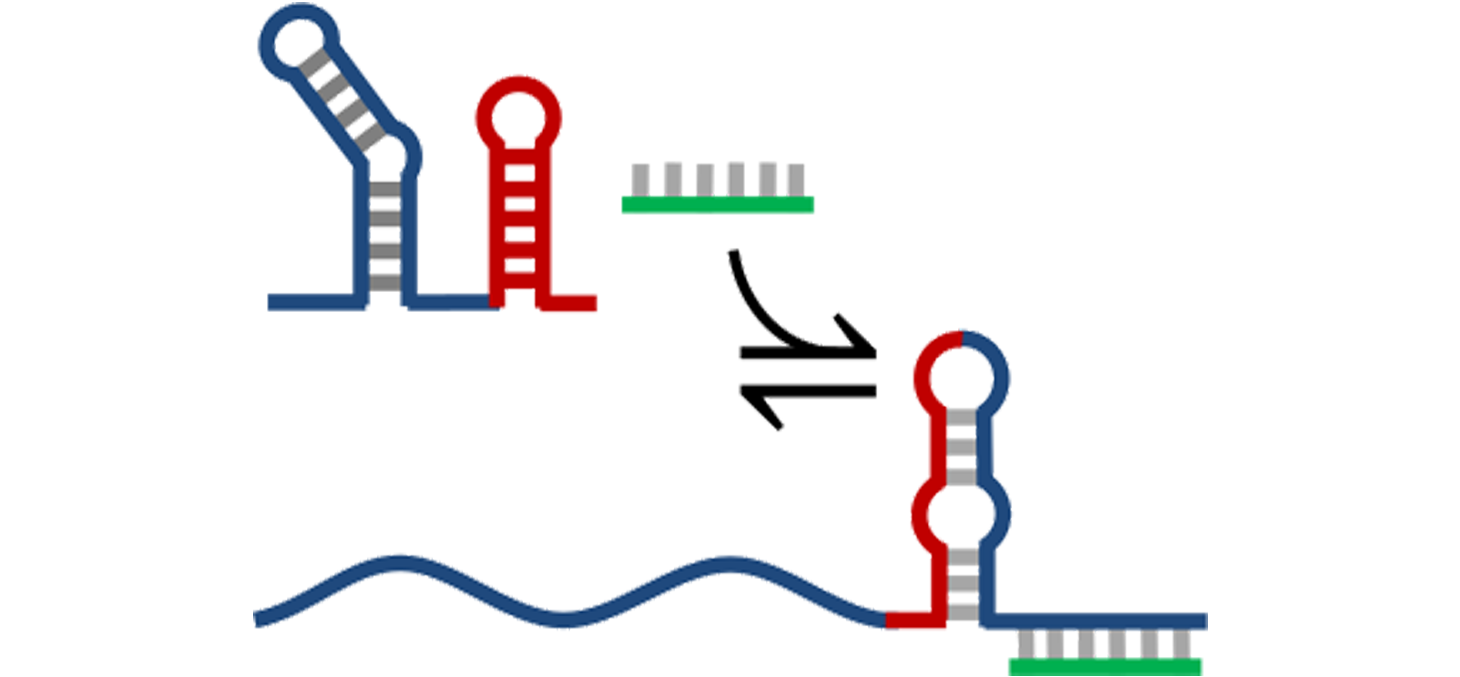
We are interested in the sequence-structure-function linkage in RNAs and in the design principles of functional RNA sequences. We address this issue through a learning-by-design approach applied to the design of synthetic RNA switches. We develop conceptual, numerical and experimental methods that take into account the dynamic nature of RNA folding.
RNA is a versatile and essential component in biological systems. An RNA sequence can fold into complex structures capable of active or structural functions in many biological processes.
Since the folding of an RNA molecule occurs as it is synthesized by RNA polymerase, RNA folding is inherently a dynamic and hierarchical process. In particular, the final structure formed results from folding intermediates and rearrangements whose order of appearance is itself determined by the sequence carried by the molecule. However, the impact of this "co-transcriptional folding" process is rarely considered in the analysis or design of functional RNA sequences.
How is the folding path that guides an RNA molecule to its final functional structure encoded in its sequence? We address this fundamental question of sequence-folding-function linkage in RNAs using a learning-by-design approach. We study the principles of encoding folding information in RNA sequences by designing functional RNA sequences and taking into account the dynamics of co-transcriptional folding to which they are naturally subjected. We apply this approach to the design of synthetic RNA switches.
- Share
- Share on Facebook
- Share on X
- Share on LinkedIn