- Share
- Share on Facebook
- Share on X
- Share on LinkedIn
Development and analysis of individual-centered models for the study of microbial growth variability
Aline Marguet, Eugenio Cinquemani
Non-permanent: Claudia Fonte Sanchez (postdoctorante), Eugène Ferragu (doctorant)
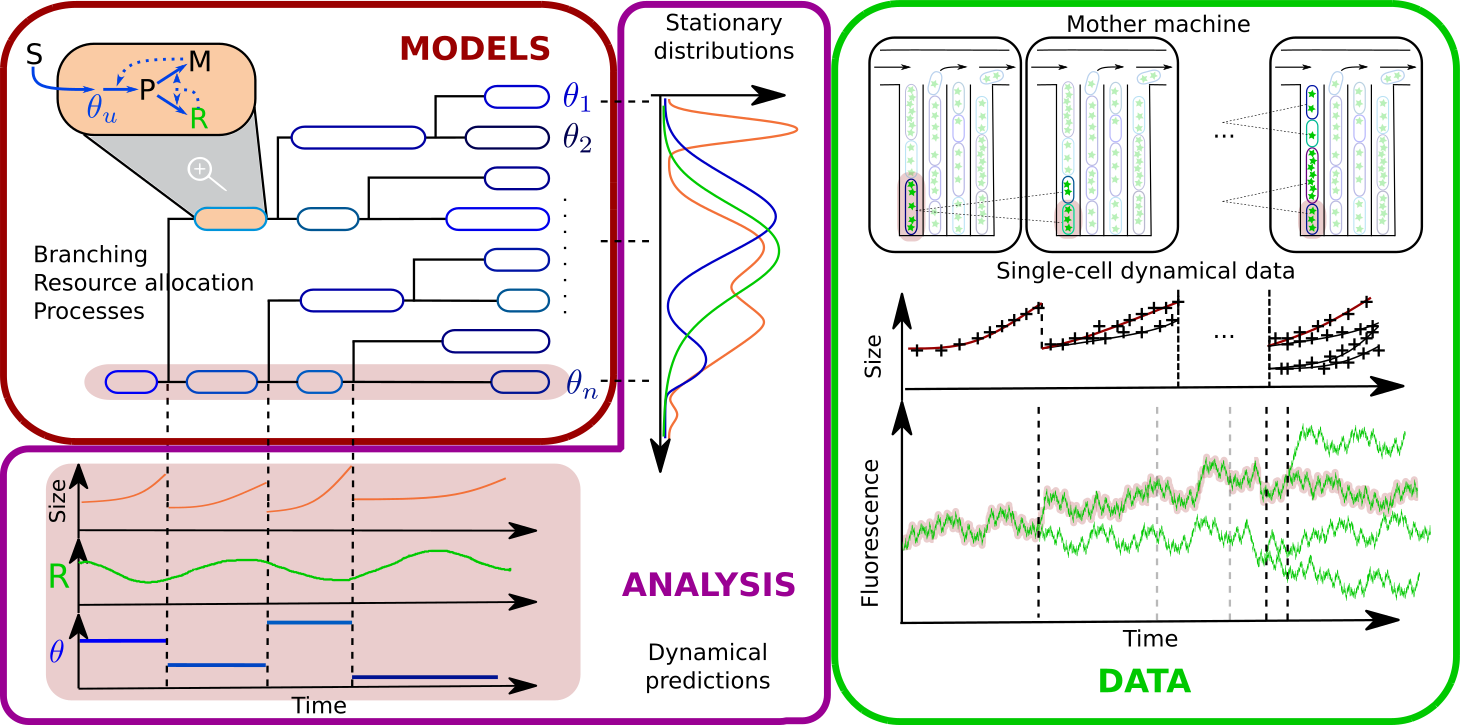
The investigation of cellular populations at a single-cell level has already led to the discovery of important phenomena, such as the occurrence of different phenotypes in an isogenic population. Nowadays, a variety of experimental techniques provide time-course measurements of growth and gene expression at the single-cell level, whether in terms of population statistics (flow cytometry) or in terms of individual cells of a lineage tree (video-microscopy combined with the use of microfluidic devices). This wealth of data enables one to take investigation of phenotypic variability at a quantitative level, provided the development of mathematical models with single-cell resolution. In particular, the development of models that take into account single-cell dynamics and genealogy is an important step in the study of the onset of variability.
Using a variety of mathematical tools, such as structured branching processes or mixed-effect models, we explore the origins of cellular variability, with focus on inheritance and stochasticity at cell division and of variability of resource allocation strategies within and across individual cells.
- Share
- Share on Facebook
- Share on X
- Share on LinkedIn