- Share
- Share on Facebook
- Share on X
- Share on LinkedIn
Publication / Research
On January 27, 2026
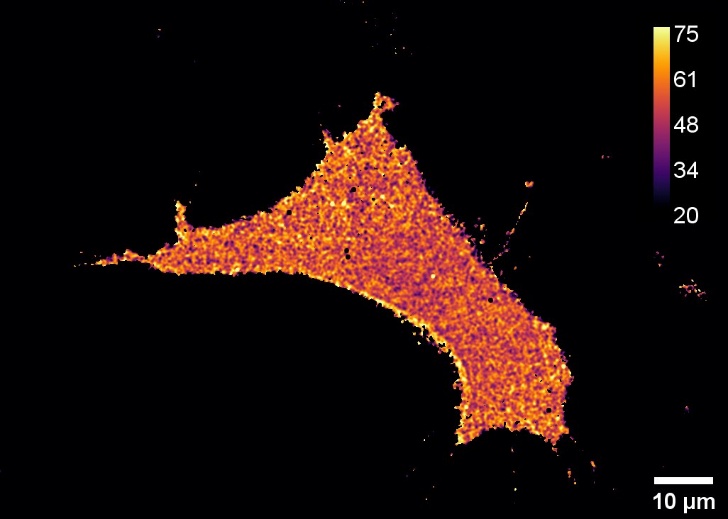
The use of fluorescent biosensors capable of optically reporting biochemical signals is hindered by the lack of simple methods to quantitatively measure and analyze their signal. The previously developed QuanTI-FRET method has been simplified and is now available in user-friendly, open-source software.
Measuring the signal reported by fluorescent biosensors based on the Förster Resonance Energy Transfer (FRET) principle is complex using a standard fluorescence microscope. Indeed, it requires measuring fluorescence intensity across three different channels and correcting the inherent biases of this technique. To address this, the QuanTI-FRET method proposed a protocol to calibrate the entire experimental setup and demonstrated absolute FRET signal measurement [1]. This calibration relied on FRET standards, whose preparation proved to be potentially cumbersome.
We propose to leverage the intrinsic properties of these biosensors to calibrate directly from the images of interest, thereby avoiding the additional step involving FRET standards. The application of this self-calibration concept is demonstrated in a scientific article [2]. To enable all research teams to benefit from this simplified yet still quantitative method, we have developed a publicly available Python analysis code [3] and also integrated it as a plugin for the open-source microscopy software Napari [4]. The method is thus usable without any knowledge of Python, simplifying both the experimental protocol and the analysis of FRET biosensors while maintaining unbiased quantitative results.
More information on the dedicated webpage.
[3] https://gricad-gitlab.univ-grenoble-alpes.fr/liphy/quanti-fret
[4] https://napari-hub.org/plugins/quanti-fret.html
Date
Contact
Aurélie DUPONT
aurelie.dupont univ-grenoble-alpes.fr (aurelie[dot]dupont[at]univ-grenoble-alpes[dot]fr)
univ-grenoble-alpes.fr (aurelie[dot]dupont[at]univ-grenoble-alpes[dot]fr)
- Share
- Share on Facebook
- Share on X
- Share on LinkedIn