- Share
- Share on Facebook
- Share on X
- Share on LinkedIn
Recruitment
Funded thesis offer 2025-2028
Supervision: Hidde de Jong and Noël Scaramozzino
Context
The PhD project will be carried out in the project-team MICROCOSME at the Centre Inria de l’Université de Grenoble Alpes and the Laboratoire Interdisciplinaire de Physique (LIPhy, CNRS/UGA) under the joint supervision of Hidde de Jong and Noël Scaramozzino within the framework of the project MuSiHC supported by the PEPR B-BEST. MICROCOSME and LIPhy provide an interdisciplinary research environment fostering close collaborative interactions between applied mathematicians, microbiologists, computer scientists, control engineers, and biophysicists.
Objectives
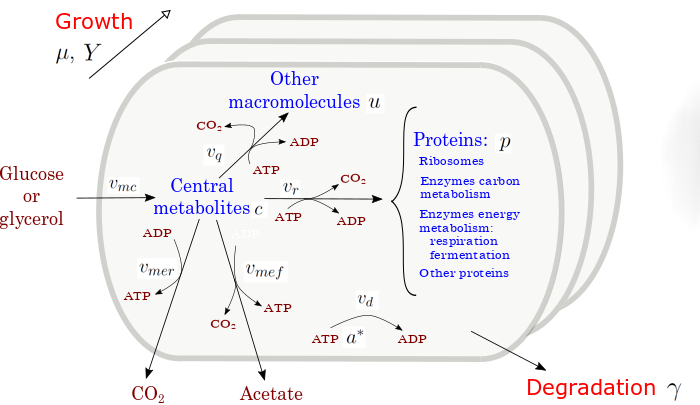
The project MUlti-SIze Hybrid Cell Models (MuSiHC) aims at developing novel hybrid approaches to the modeling of cells and bioreactors for the production of added-value compounds. In particular, the project will develop a toolkit of hybrid models of different sizes, combining a mechanistic description with AI/ML components [1], to obtain more reliable cell and bioreactor simulations. As a proof of concept, the project will focus on Escherichia coli as a platform for the bioproduction of 1,3-propanediol (1,3-PDO), a high-value compound with vast applications in the chemical industry.
The proposed PhD project is concerned with the development of a small-scale, dynamic models [2,3] for optimizing the production of 1,3-PDO by E. coli, involving such tasks as model formulation and reduction, running mini-bioreactor experiments for model calibration, using the models to identify conditions for optimal metabolite production, and the experimental test of these conditions. The PhD project involves active collaboration with other MuSiHC partners at INRAE (Jean-Loup Faulon, Wolfram Liebermeister) and Toulouse Biotechnology Institute (César Arturo Aceves Lara). Beyond the specific application of MuSiHC, the project aims at identifying general principles for the development and validation of small-scale models of biotechnological production systems.
[1] Faure, L., Mollet, B., Liebermeister, W., & Faulon, J. L. (2023). A neural-mechanistic hybrid approach improving the predictive power of genome-scale metabolic models. Nature Communications, 14(1):4669. https://doi.org/10.1038/s41467-023-40380-0
[2] Baldazzi, V., Ropers, D., Gouzé, J. L., Gedeon, T., & de Jong, H. (2023). Resource allocation accounts for the large variability of rate-yield phenotypes across bacterial strains. eLife, 12:e79815. https://doi.org/10.7554/eLife.79815
[3] Wortel, M. T., Noor, E., Ferris, M., Bruggeman, F. J., & Liebermeister, W. (2018). Metabolic enzyme cost explains variable trade-offs between microbial growth rate and yield. PLoS Computational Biology, 14(2):e1006010. https://doi.org/10.1371/journal.pcbi.1006010
Main activities
The PhD project is an interdisciplinary project involving both the development of mathematical models describing the biological system and experimental work to calibrate and validate the models:
- Reduction of a medium-scale, kinetic model to a small-scale, whole-cell model of the production of 1,3-propanediol (1,3-PDO) by Escherichia coli, using previously developed reduction methods and taking inspiration from existing small-scale resource allocation models.
- Performance of experiments with selected E. coli strains on an in-house mini-bioreactor platform to obtain data (growth, gene expression, metabolite concentrations) for the calibration of the model.
- Use a combination of optimization and simulation approaches to identify conditions maximizing 1,3-PDO production.
- Validation of the predicted optimal operating conditions by performing the corresponding mini-bioreactor experiments, including the quantification of 1,3-PDO production.
Requirements
Interested candidates are ideally expected to have some experience with the mathematical modelling of biological systems and/or laboratory work in microbiology, but we are open to consider students with good scholarly results motivated by interdisciplinary research from a range of fields (microbiology, mathematical biology, bioinformatics, ecology, biophysics, …).
Starting date
Fall 2025
Contact and applications
If you are interested to learn more about the position, please contact Hidde de Jong (hidde.de-jong inria.fr (hidde[dot]de-jong[at]inria[dot]fr)) or Noël Scaramozzino (natale.scaramozzino
inria.fr (hidde[dot]de-jong[at]inria[dot]fr)) or Noël Scaramozzino (natale.scaramozzino univ-grenoble-alpes.fr (natale[dot]scaramozzino[at]univ-grenoble-alpes[dot]fr)).
univ-grenoble-alpes.fr (natale[dot]scaramozzino[at]univ-grenoble-alpes[dot]fr)).
Download
Contact
Hidde de Jong
BIOP team
hidde.de-jong inria.fr (hidde[dot]de-jong[at]inria[dot]fr)
inria.fr (hidde[dot]de-jong[at]inria[dot]fr)
Noël Scaramozzino
BIOP team
natale.scaramozzino univ-grenoble-alpes.fr (natale[dot]scaramozzino[at]univ-grenoble-alpes[dot]fr)
univ-grenoble-alpes.fr (natale[dot]scaramozzino[at]univ-grenoble-alpes[dot]fr)
- Share
- Share on Facebook
- Share on X
- Share on LinkedIn