- Imprimer
- Partager
- Partager sur Facebook
- Partager sur X
- Partager sur LinkedIn
Séminaire
Le 13 mars 2025
Marc Joyeux (PSM, LIPhy)
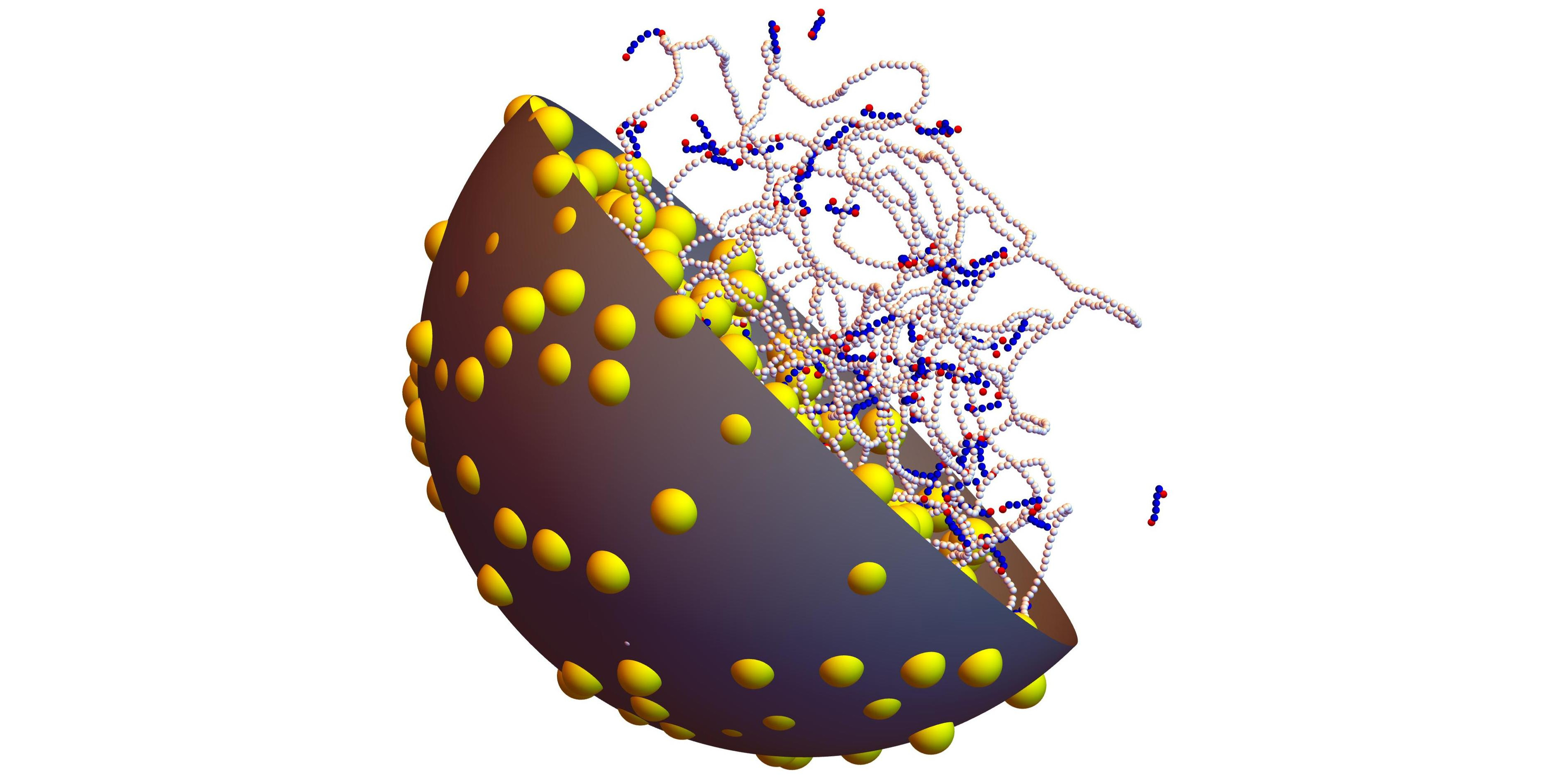
The nucleoid, that is the region of bacterial cells that contains the genomic DNA as well as several tens of thousands of associated proteins, is rather interesting from a biophysical point of view, because its formation, organization and dynamics rely not only on specific chemical interactions, like all biological processes, but also on a few general features pertaining essentially to polymer physics. Elucidating the role and importance of these mechanisms is a rather exciting challenge.
Another interesting point concerning the bacterial nucleoid is that even fundamental observations, like the 1000-fold compaction of the DNA inside the nucleoid and its separation from the rest of the cytoplasm, lack a definitive explanation and are still the subject of lively debates. In the present talk, I will precisely discuss recent work showing how coarse-grained modeling (at the level of 1 small particle per 8 DNA base pairs and 1 bigger particle for macromolecules of the size of ribosomes) and Brownian Dynamics simulations help understand the dynamics of the nucleoid [Organization of the bacterial nucleoid by DNA-bridging proteins and globular crowders, Frontiers in Microbiology 14, 1116776 (2023)].
The two most important contributions to the compaction of the DNA coil are arguably the cross-linking of the DNA by nucleoid proteins (like H-NS and StpA), which corresponds to an associative phase separation, and the demixing of DNA and other abundant globular macromolecules which do not bind to the DNA (like ribosomes), which corresponds to a segregative phase separation. Brownian Dynamics simulations indicate that the radius of gyration of the DNA coil decreases linearly with the effective volume ratio of globular crowders and the number of DNA bridges formed by nucleoid proteins in the whole range of physiological values. They also highlight the fact that the number of DNA bridges formed by nucleoid proteins depends crucially on their ability to self-associate (oligomerize). An explanation for this result is proposed in terms of the mean distance between DNA segments and the capacity of proteins to maintain DNA–bridging in spite of the thermal fluctuations of the DNA network. Finally, simulations indicate that non-associating proteins preserve a high mobility inside the nucleoid, leading to a DNA/protein complex which looks like a liquid droplet. In contrast, self-associating proteins form a little deformable network which cross-links the DNA chain, with the consequence that the DNA/protein complex looks more like a gel.
Date
11:00
Localisation
LIPhy, salle de conférence
- Imprimer
- Partager
- Partager sur Facebook
- Partager sur X
- Partager sur LinkedIn