- Share
- Share on Facebook
- Share on X
- Share on LinkedIn
Séminaire
On December 19, 2023
Marta Bally (Department of Clinical Microbiology & Wallenberg Centre for Molecular Medicine, Umeå University, Umeå, Sweden)
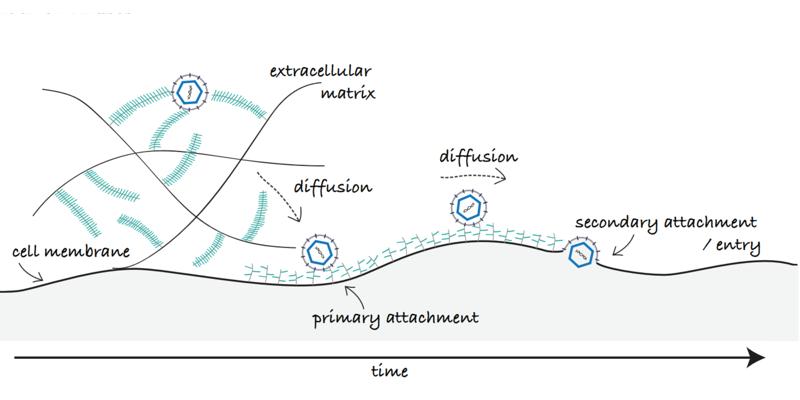
Viruses are small pathogenic particle that rely on hijacking a cellular host to replicate and spread. The initial recruitment of a virus particle to the cell surface is a complex dynamic multistep process that requires diffusion of the virus particle through the glycocalyx, the sugar coat covering cells, to reach the cell membrane where it may further diffuse laterally in search of a suitable point of entry. Many viruses, initiate their recruitment on the host cell by binding to carbohydrates found on the cell surface and in the glycocalyx, sulfated glycosaminoglycans (GAGs) such as heparan sulfate, for example. This initial recognition is crucial in the viruses’ life cycle as it leads to infection. Equally important is however, the capability of the virus to overcome these interactions upon egress to ensure virus propagation. A tight regulation of such interactions is also essential in the context of virus transport at the cell surface; binding a high amount of receptors with high affinity might trap the virion at the cell surface before it meets entry receptors, while a too weak binding may not be enough for attachment of the particle or to ensure sufficient residence time for entry.
In our work, we study the molecular mechanisms modulating virus attachment, detachment and diffusion at the cell surface. We primarily focus on virus-GAG interactions involved amongst other in the initial recruitment of herpes simplex virus (HSV), human papilloma virus (HPV) and severe and acute respiratory syndrome coronavirus-2 (SARS-CoV-2). Our experimental approach is based on probing interactions between individual virus particles and the cell surface using both cell-surface mimics of controlled biomolecular composition, and live cell microscopy. Single particle tracking and total internal reflection fluorescence microscopy are used to study the interaction kinetics and diffusion of individual viruses. Complementary atomic force microscopy-based single molecule force spectroscopy contributes to studying individual ligand-receptor interactions involved in the process.
Taken together, our research contributes to a better understanding of the mechanisms regulating the interaction between a virus and the surface of its host. Such insights will without doubt facilitate the design of more efficient antiviral drugs or vaccines.
[1] Altgarde, N., et al., Mucin-like Region of Herpes Simplex Virus Type 1 Attachment Protein Glycoprotein C (gC) Modulates the Virus-Glycosaminoglycan Interaction. Journal of Biological Chemistry, 2015. 290(35): p. 21473-21485.
[2]. Peerboom, N., et al., Binding Kinetics and Lateral Mobility of HSV-1 on End-Grafted Sulfated Glycosaminoglycans. Biophysical Journal, 2017. 113(6): p. 1223-1234.
[3] Delguste et al. Regulatory Mechanisms of the Mucin-Like Region on Herpes Simplex Virus during Cellular Attachment. ACS Chemical Biology, 2019, 14, 3, 534–542
[4] Abidine, Y. et al.,Cellular glycosaminoglycans and viral carbohydrates regulate diffusion of herpesvirus simplex virus 1 in the glycocalyx. Viruses, 2022, 14, 8, 1836.
[5] Bally, M. et al., Physicochemical tools for studying virus interactions with targeted cell membranes in a molecular and spatiotemporally resolved context. Anal Bioanal Chem, 2021, 413, 7157–7178
Contact: Delphine Débarre
Date
14:00
Localisation
LIPhy, salle de conférence
- Share
- Share on Facebook
- Share on X
- Share on LinkedIn